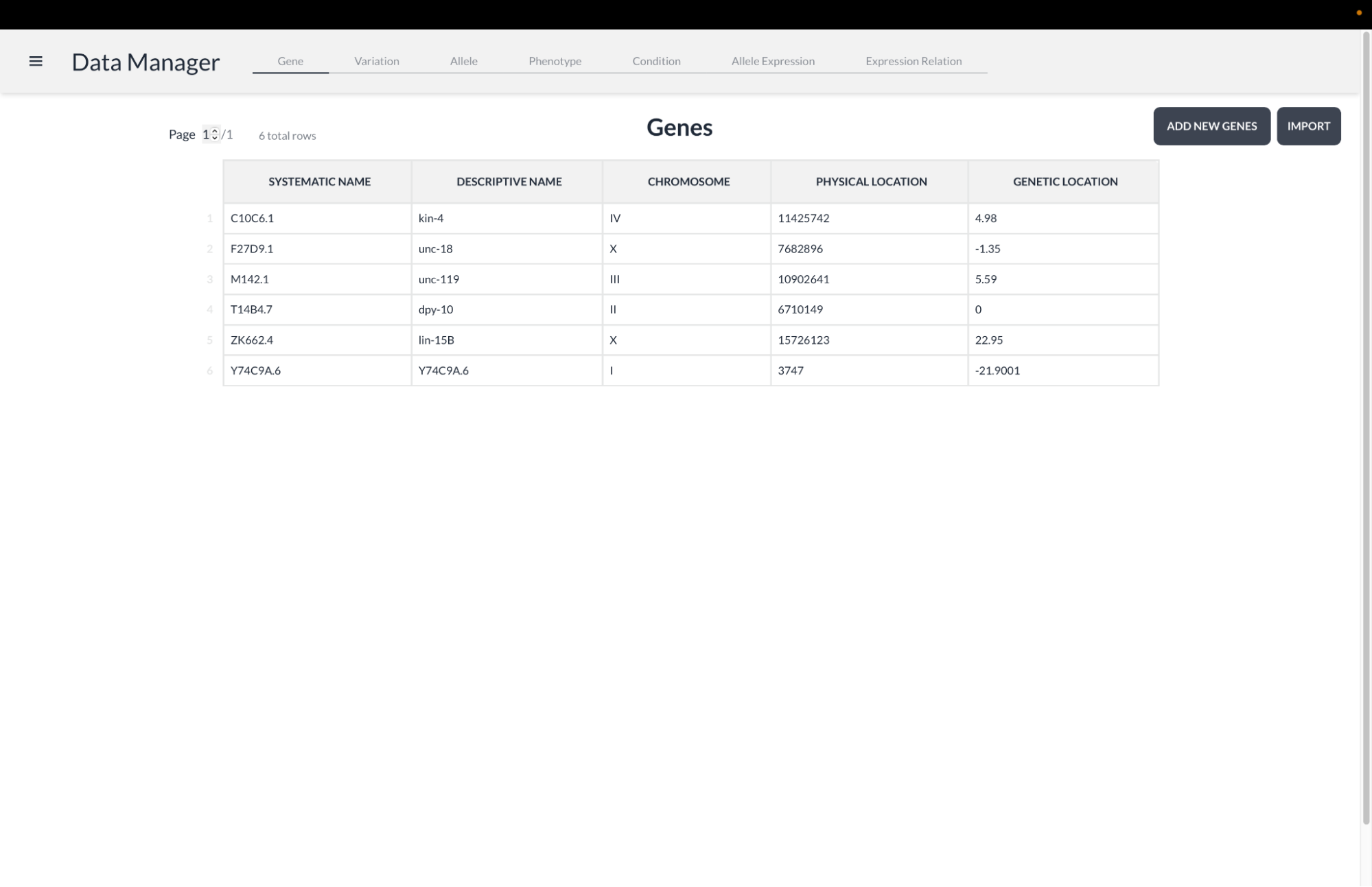
WormWorld is a multi-platform desktop application to assist modern genetic researchers in a laboratory setting. It specifically supports those who study the model organism C. Elegans, which is a small roundworm whose genetics are commonly studied in labs around the world to provide insights into basic genetics and analogous genetic processes in humans. We are sponsored by Matt Rich and Wayne Davis of the University of Utah School of Biological Sciences.
Genetics is a large and complex field, and a cursory search for genetics software shows an abundance of tools to match the wide variety of areas, approaches, and research methods used by geneticists. By its nature, genetics lends itself to computational assistance because of the kinds and quantities of data that can be analyzed and processed; it is not surprising that in their attempts to uncover the mysteries of genetic code, researchers and developers have turned to computer code and the computational power and productivity enhancements it can proffer. One of our sponsors, in fact, has developed and published a tool called ApE, which is a “plasmid editor.” One of the most common laboratory activities is to plan and execute a sequence of related genetic cross-breedings or “crosses” to arrive at a desired genetic profile. Despite this, there does not seem to be an existing well-known gene-crossing designer or scheduler to facilitate these activities. Consequently, these designs and schedules must be done by hand, which can be time consuming and error prone. As a solution to this, WormWorld provides tools for designing and scheduling genetic crosses. It includes technical conveniences such as automatic offspring calculations with statistical frequencies as well as practical and logistical features to organize and coordinate the execution of genetic crosses.
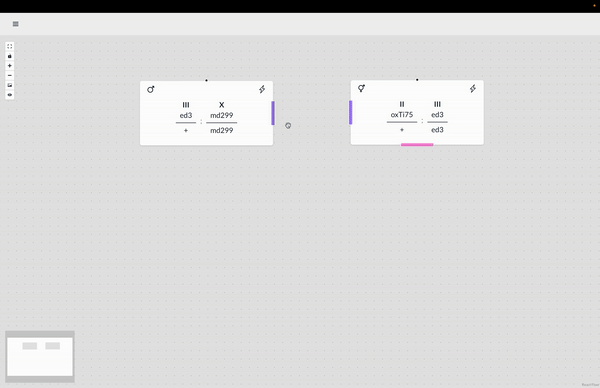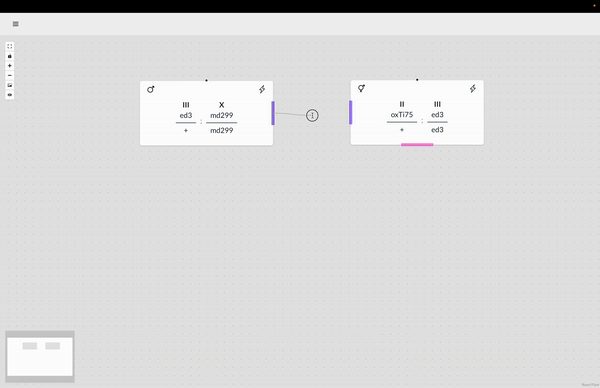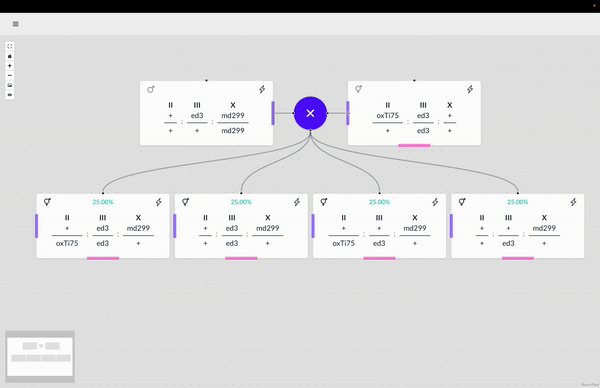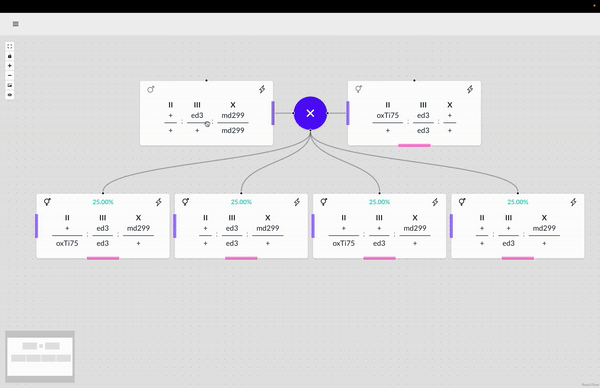
The WormWorld application promises to fill an open niche in this area and potentially enhance genetic-crossing research productivity. Beyond this principal use, there are secondary uses for the application–as a pedagogical tool, for example, that can help illustrate the nature of genetic crosses or bridge the experience gap for a new researcher through an interactive design process and the display of helpful information.
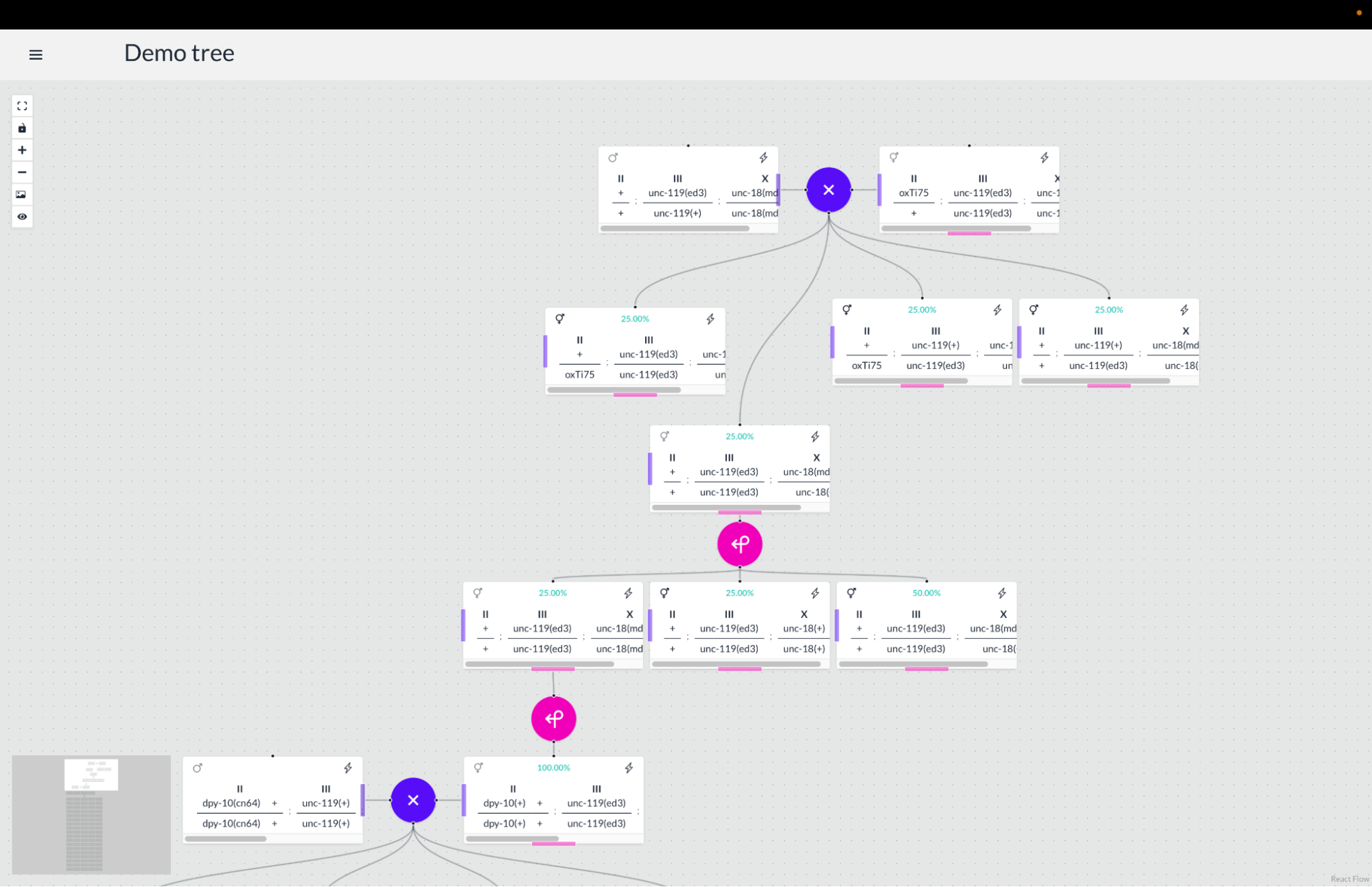
Wormworld is built using the Tauri app framework, which allowed us to develop the frontend with web technologies and the backend with the Rust language. Tauri is an alternative to Electron that helps developers build lean and performant apps. For the frontend, we use React with Typescript to achieve reactive user interfaces with type safety. We style the application with TailwindCSS, which provides utility classes for consistent and attractive styling, as well as DaisyUI, which provides standardized TailwindCSS component styling. For the backend, we use Sqlite3 bindings for Rust to store and query the data for strains, genes, alleles, and breeding timelines. For both the front and backend, we have compile-time type checking and extensive testing to help assure consistent and robust software. The app runs on a user’s desktop with a local Sqlite3 database that they can import and export data from.
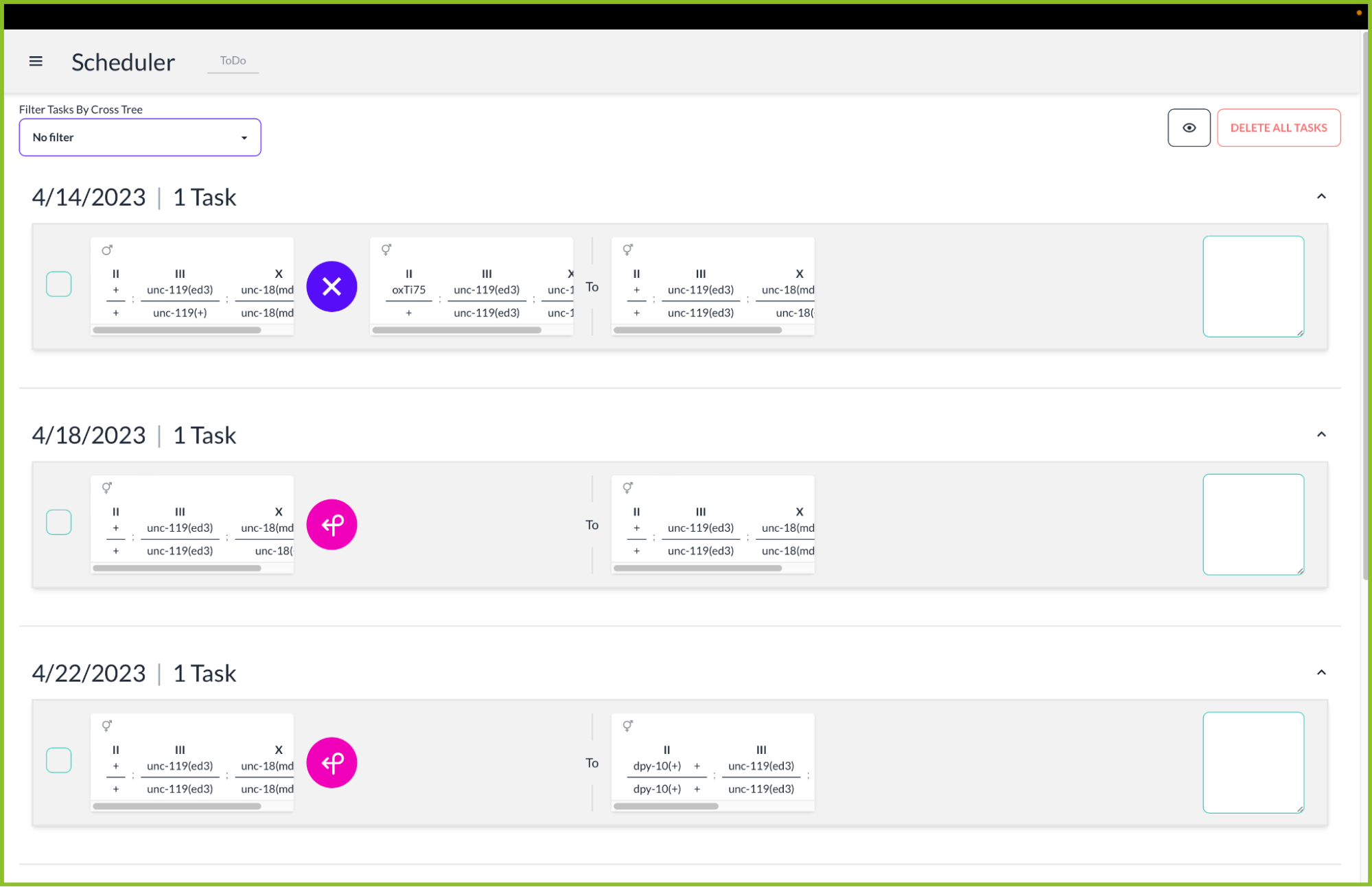
The Tauri app framework uses each operating system’s native webviews to build cross-platform desktop applications. This allows our app to run on Linux, MacOS, and Windows. If a desktop computer can run a browser, it can run our application. The installation process for a user is simple: download the installer on Windows, a bundle on MacOS, or a .deb package for Linux. Tauri includes an updater in the binaries, so once a user has installed our app, we can publish updates to them without requiring them to manually download it again.